Read and download free pdf of CBSE Class 12 Biology Biotechnology Principles And Processes Worksheet Set B. Students and teachers of Class 12 Biology can get free printable Worksheets for Class 12 Biology Chapter 9 Biotechnology Principles and Processes in PDF format prepared as per the latest syllabus and examination pattern in your schools. Class 12 students should practice questions and answers given here for Biology in Class 12 which will help them to improve your knowledge of all important chapters and its topics. Students should also download free pdf of Class 12 Biology Worksheets prepared by teachers as per the latest Biology books and syllabus issued this academic year and solve important problems with solutions on daily basis to get more score in school exams and tests
Worksheet for Class 12 Biology Chapter 9 Biotechnology Principles and Processes
Class 12 Biology students should download to the following Chapter 9 Biotechnology Principles and Processes Class 12 worksheet in PDF. This test paper with questions and answers for Class 12 will be very useful for exams and help you to score good marks
Class 12 Biology Worksheet for Chapter 9 Biotechnology Principles and Processes
Question. Small chemically synthesised oligonucleotides that are complementary to the regions of DNA at 3' end used in PCR are :
(a) Primers
(b) Dimers
(c) Small strands
(d) Large fragments
Answer : A
Question. Which one of the following is must in Biotechnology?
(a) Restriction endonuclease + DNA ligase
(b) Restriction exonuclease + DNA polymerase
(c) Alkaline phosphate + DNA Ligase
(d) RNA polymerase + DNA polymerase
Answer : A
Question. Taq. polymerase is obtaned from :
(a) Bacillus thuriengiensis
(b) Thermus aquaticus
(c) Salmonella typhimurium
(d) Eischerichia coli
Answer : B
Question : The first human hormone produced by recombinant DNA technology is :
(a) Insulin
(b) Estrogen
(c) Thyroxine
(d) Progesterone
Answer : A
Question : Transgenic animals are
(a) Animals whose DNA is manipulated to possess and express an extra (foreign) gene.
(b) Animals whose RNA is manipulated to possess and express an extra (foreign) gene.
(c) Animals whose both DNA and RNA are manipulated to possess and express an extra (foreign) gene.
(d) None of the above.
Answer : A
Question : Which of the following is commonly used as a vector for introducing a DNA fragment in human lymphocytes ?
(a) Retrovirus
(b) pBR 322
(c) l Phage
(d) Ti plasmid
Answer : A
Question : Maximum number of existing transgenic animals is of
(a) Mice
(b) Cow
(c) Pig
(d) Fish
Answer : A
Question : GEAC stands for
(a) Genome Engineering Action Committee.
(b) Ground Environment Action Committee.
(c) Genetic Engineering Approval Committee.
(d) Genetic and Environment Approval committee.
Answer : C
Question : Crystals of Bt toxin produced by some bacteria do not kill the bacteria themselves because
(a) bacteria are resistant to the toxin.
(b) toxin is immature.
(c) toxin is inactive.
(d) bacteria enclose toxin in a special sac.
Answer : C
Question : Choose the correct option regarding Retrovirus
(a) RNA virus that can synthesise DNA during infection.
(b) a DNA virus that can synthesise RNA during infection.
(c) a ss DNA virus.
(d) a dsRNA virus.
Answer : A
Question : India has……. varieties of Basmati rice.
(a) 30
(b) 29
(c) 12
(d) 27
Answer : D
Question : ………….. is a first transgenic cow.
(a) Dolly
(b) Molly
(c) Shelly
(d) Rosie
Answer : D
Question : Use of bio-resources by multinational companies and organisations without authorisation from the concerned country and its people is called
(a) Bio-infringement
(b) Bioexploitation
(c) Biodegradation
(d) Biopiracy
Answer : D
Question. Which of the following is used to deliver desirable gene in to animal cell :
(a) Disarmed retrovirus
(b) Disarmed agrobacterium
(c) Disarmed E.coli
(d) Disarmed plant pathogen
Answer : A
Question. Agrobacterium tumifaciencs, a pathogen of several dicot plants is able to deliver a piece of DNA and it is known as :
(a) R-DNA
(b) S-DNA
(c) M-DNA
(d) T-DNA
Answer : D
Question. To denature the DNA template in PCR it is heated to
(a) 70°C
(b) 54°C
(c) 80°C
(d) 94°C
Answer : D
Question. Roman numbers following the names of restriction endonuclease indicate :
(a) The order in which the enzymes were isolated from that strain of bacteria
(b) strain of bacteria
(c) the order in which genus is taken to isolate the enzyme
(d) none of the above
Answer : A
Question. The science, which deals with techniques of using live organisms or enzymes from organism to produce products and processes useful to human is :
(a) Genetics
(b) Biotechnology
(c) Bioinformatics
(d) None of these
Answer : B
Question. Autonomously replicating circular extra chromosomal DNA of bacteria is :
(a) Plastid
(b) Nucleus
(c) Plasmid
(d) None of these
Answer : C
Question. Exonuclease removes nucleotides from
(a) Specific positions
(b) the ends of the DNA
(c) any where in DNA
(d) All the above
Answer : B
Question. Alternative selectable markers developed to differentiate non-recombinants from recombinants on the basis of :
(a) Ability of separate them according to size
(b) Ability to produce colour in the presence of a chromogenic substrate
(c) Ability to not produce colour
(d) None of the above
Answer : B
Question. A restriction endonucleases which always cut DNA molecules at a particular point by recognising a specific sequence of six base pairs is :
(a) Hind-II
(b) Psu I
(c) Hae-III
(d) All of these
Answer : A
Question. The first letter of the name of Restriction endonuclease came from the
(a) Genus of organism
(b) Species of organism
(c) Family of organism
(d) Class of organism
Answer : A
Question. Father of genetic engineering is :
(a) Paul Berg
(b) Nathans
(c) Herbert Boyer
(d) Stanley Cohen
Answer : A
Question. A definition of biotechnology that encompasses both traditional view and modern view are given by :
(a) European forum on Biotechnology
(b) European focus on Biotechnology
(c) European Federation of Biotechnology
(d) European Centre of Biotechnology
Answer : C
Question. In a chromosome there is a specific DNA sequence which is responsible for initiating replication is :
(a) Ori
(b) Palindromic sequence
(b) Initiation sequence
(d) Promoter sequence
Answer : A
Question. To make cell competent to take up DNA, heat shock is given to cells, the temperature of shock is :
(a) 30°C
(b) 42°C
(c) 60°C
(d) 90°C
Answer : B
Question. In gel electrophoresis technique the DNA fragments are forced to move through a medium towards :
(a) Anode
(b) Cathode
(c) Both (a) and (b)
(d) None of the above
Answer : A
Question. First recombinant DNA was made by Stanley Cohen and Herbert Boyer in :
(a) 1968
(b) 1970
(c) 1972
(d) 1974
Answer : C
Question. The first restriction endonuclease discovered, was
(a) Eco RI
(b) Sam I
(c) Bam HI
(d) Hind II
Answer : D
Question. In the vector pBR322 there is
(a) One selectable marker
(b) Two selectable markers
(c) Three selectable markers
(d) None of the above
Answer : B
Question. When the isolation of genetic material is done the RNA can be removed by treatment with :
(a) Protease
(b) Chitinase
(c) Ribonuclease
(d) Deoxyribonuclease
Answer : C
Question. DNA fragments generated by the restriction endonucleases in a chemical reaction can be separated by
(a) polymerase chain reaction
(b) electrophoresis
(c) restriction mapping
(d) centrifugation
Answer : B
Question. Reverse transcription PCR uses _________.
(a) RNA as a template to form DNA
(b) mRNA as a template to form cDNA
(c) DNA as a template to form ssDNA
(d) All of the above
Answer: B
Question. Which of the following is not a feature of the plasmid?
(a) Single stranded
(b) Independent replication
(c) Circular structure
(d) Small, circular double-stranded
Answer: A
Question. Continuous culture has cells in
(a) their physically active phase
(b) Lag phase
(c) Exponential phase
(d) Both a and b
Answer: C
Question. How many DNA duplexes are obtained from one DNA duplex after 4 cycles of PCR?
(a) 8
(b) 4
(c) 32
(d) 16
Answer: D
Question. The construction of the first recombinant DNA was by linking of a gene encoding antibiotic resistance with the native plasmid of
(a) Salmonella typhimurium
(b) Agrobacterium tumefaciens
(c) Entamoeba histolytica
(d) Streptococcus pneumoniae
Answer: A
Question. Identify a character that is not desirable in a cloning vector
(a) an inactive promoter
(b) an origin of replication site
(c) selectable markers such as genes for antibiotic resistance
(d) one or more unique restriction endonuclease site
Answer: A
Question. The cylindrical or curved base of a bioreactor actually facilitates:
(a) The handling and maintenance of bioreactor.
(b) Mixing
(c) Better oxygen transport
(d) More accumulation of product
Answer: B
Question. Which of the following statements is accurate for the PCR – polymerase chain reaction?
(a) Automated PCR machines are called thermal cyclers
(b) A thermostable DNA polymerase is required
(c) Millions to billions of desired DNA copies can be produced from microgram quantities of DNA
(d) All of the above
Answer: D
Question. A cloning vector has two antibiotic resistance genes- for tetracycline and ampicillin. A foreign DNA was inserted into the tetracycline gene. Nonrecombinants would survive on the medium containing.
(a) ampicillin but not tetracycline
(b) tetracycline but not ampicillin
(c) both tetracycline and ampicillin
(d) neither tetracycline nor ampicillin
Answer: C
Question. Primers used for the process of polymerase chain reaction are _________.
(a) Single-stranded RNA oligonucleotide
(b) Single-stranded DNA oligonucleotide
(c) Double-stranded RNA oligonucleotide
(d) Single-stranded DNA oligonucleotide
Answer: B
Question. Which of the following restriction sites is located within the gene for tetracycline resistance in the plasmid pBR322?
(a) BamHI
(b) Psti
(c) Clal
(d) Pvull
Answer: A
Question. Plasmid has been used as vector because
(a) both its ends show replication.
(b) it can move between prokaryotic and eukaryotic cells.
(c) it is circular DNA which have capacity to join to eukaryotic DNA.
(d) it has antibiotic resistance gene.
Answer: C
Question. Which of the following is not a component of downstream processing?
(a) Expression
(b) Preservation
(c) Purification
(d) Separation
Answer: A
Question. The EcoR-1 enzyme is obtained from
(a) Virus
(b) Salmonella
(c) E.Coli
(d) Penicillium
Answer: C
Question. A and B in the pBR 322, shown in the diagram given below, respectively represent recognition sequences of (19)
(a) BamH I and Sma I
(b) Hind II and Sma I
(c) BamH I and SaI I
(d) SaI I and Hind II
Answer: C
Question. A gene, whose expression helps to identify transformed cells is known as
(a) selectable marker
(b) vector
(c) plasmid
(d) structural gene
Answer: A
Question. A foreign DNA and plasmid cut by the restriction endonuclease can be joined to form a recombinant plasmid using_________
(a) Eco RI
(b) Taq polymerase
(c) Ligase
(d) Polymerase II
Answer: C
Question. The DNA molecule to which the gene of interest is integrated for cloning is called
(a) Transformer
(b) Vector
(c) Template
(d) Carrier
Answer: B
Question. In agarose gel electrophoresis, DNA molecules are separated on the basis of their
(a) Size only
(b) Charge to size ratio
(c) All of the above
(d) Charge only
Answer: A
Question. The linking of antibiotic resistance gene with the plasmid vector became possible with
(d) ampR, tetR-antibiotic resistance genes
(b) endonucleases
(c) DNA polymerase
(d) exonucleases
Answer: B
Question. Microinjection is suitable for
(a) Injecting an ovum into the sperm in IVF
(b) Transforming animal cells
(c) Injecting very small sized drug particles into neurons
(d) Conferring antibiotic resistance to a certain strain of bacteria
Answer: B
Question. The plasmid pBR322 does not contain.
(a) An origin of replication site
(b) A gene that encodes for restrictor of plasmid copy number
(c) Gene for Ampicillin and Streptomycin resistance
(d) Sites for many restriction enzymes
Answer: B
Question. Plasmid vector in DNA recombinant technology means
(a) a virus that transfers gene to bacteria
(b) extra-chromosomal autonomously replicating circular DNA
(c) sticky end of DNA
(d) any fragment of DNA carrying desirable gene
Answer: B
Question. Which organism is used to transfer T-DNA?
(a) Streplomyceshy groscopicus
(b) Agrobacterium tumefaciens
(c) Salmonella typhi
(d) Escherichia coli
Answer: B
Question. The technique not used for transformation of plant cells in recombinant procedures is:
(a) Biolistic
(b) Agrobacterium mediation
(c) Use of viruses
(d) Micro-injection
Answer: B
Question. What does continuous culture mean?
(a) Where DNA and protein getting expressed continuously.
(b) Where the used medium is drained out while fresh medium is added from another side.
(c) Where cells are producing protein for one week continuously.
(d) Where the production of recombinant DNA is continuing without interference.
Answer: B
Question. The first restriction endonuclease reported was
(a) Hind II
(b) EcoRI
(c) Hind III
(d) BamHI
Answer: A
Question. Which enzyme is used to isolate DNA from fungal cells for biotechnology experiments?
(a) Lysozyme
(b) Chitinase)
(c) Cellulase
(d) Ligase.
Answer: B
Question. Suggest a technique to a researcher who needs to separate fragments of DNA?
(a) PCR
(b) Gel electrophoresis.
(c) Centrifugation
(d) X-ray diffraction.
Answer: B
Question. If DNA is inserted within the coding sequence of b-galactosidase enzyme then
(a) Non-recombinants will give blue coloured colonies in presence of chromogenic substrate
(b) Recombinant will give blue coloured colonies in presence of chromogenic substrate
(c) Both recombinants and non-recombinants give blue colour
(d) Non-recombinants do not produce colour due to insertional inactivation.
Answer : A
Question. Knife of DNA :
(a) DNA - ligase
(b) Restriction endonuclease
(c) Exonuclease
(d) Peptidase
Answer : B
Question. Insertional inactivation results into inactivation of which enzyme ?
(a) Transacetylase
(b) Permease
(c) Taq polymerase
(d) b-galactosidase
Answer : D
Assertion - Reason Question:
In the following questions, a statement of assertion is followed by a statement of reason. Mark the correct choice as:
(a) If both Assertion and Reason are true and Reason is the correct explanation of Assertion.
(b) If both Assertion and Reason are true but Reason is not the correct explanation of Assertion.
(c) If Assertion is true but Reason is false.
(d) If both Assertion and Reason are false.
Question. Assertion: Genetic engineering can overcome the drawbacks of traditional hybridization.
Reason: Genetic engineering can create desired DNA sequences to meet specific requirements.
Answer: A
Question. Assertion: The process including separation & purification of product are collectively referred as downstream processing
Reason: The downstream processing & quality control testing are same for all type of products.
Answer: C
Question. Assertion: In PCR, primers are used which are single stranded RNA fragment 100-200 nucleotides in length.
Reason: The primers get attached to 5‘ site of the target sequence.
Answer: D
Question. Assertion: Restriction endonucleases are also called 'molecular scissors'.
Reason: When fragments generated by restriction endonucleases are mixed, they join together due to their sticky ends.
Answer: B
Question. Assertion: Plasmids are extra chromosomal DNA.
Reason: Plasmids are found in bacteria and are useful in genetic engineering.
Answer: B
Question. Assertion: A piece of DNA inserted into an alien organism generally does not replicate if not inserted into a chromosome.
Reason: Chromosomes have specific sequences called 'ori' region where DNA replication is initiated.
Answer: A
Question. Assertion: The stirred-tank is well suited for large scale production of microorganisms under aseptic conditions.
Reason: In sparged stirred tank bioreactor, surface area for oxygen transfer is increased.
Answer: B
Question. Assertion: To extract DNA from tomato cells, first it must be treated with chitinase, protease and then deoxyribonuclease.
Reason: Deoxyribonuclease will digest Proteins While RNA will be intact.
Answer: D
Question. Assertion: The most commonly used bioreactors are of stirring type.
Reason: The stirrer facilitates even mixing & oxygen availability throughout the bioreactor.
Answer: A
Question. Assertion: Insertion of recombinant DNA within the coding sequence of β-galactosidase results in colourless colonies.
Reason: Presence of insert results in inactivation of enzyme β- galactosidase known as insertional inactivation.
Answer: A
Very Short Answer Type Questions:
Question. An extra chromosomal segment of circular DNA of a bacterium is used to carry gene of interest into the host cell. What is the name given to it?
Answer: Plasmid.
Question. Name the enzymes used in Bacteria, Plants and fungus for isolating DNA?
Answer: Lysozyme for bacteria, cellulase for plant cell and chitinase for fungus.
Question. Make a list of tools of recombinant DNA technology.
Answer: Key tools of recombinant DNA technology:
Restriction enzymes
Polymerase enzymes
Ligases
Vectors
Host organism.
Question. Why is the primer required in PCR added at the 3‘ end of DNA template?
Answer: The dna polymerase can catalyse polymerisation reaction in 5‘ to 3‘ direction only. The free 3‘ end of the primer will provide linking site for new nucleotides.
Question. Name two commonly used vectors in genetic engineering.
Answer: Plasmid and Bacteriophage.
Question. What are the other macromolecules associated with DNA?
Answer: RNA, proteins, polysaccharides and also lipids.
Question. Identify the recognition sites in the given sequences at which E.coRI will cut and make sticky ends.
5´-GAATTC-3´
3´-CTTAAG-5´
Answer: 5‟−G↓AATTC-3‟
3‟−CTTAA↑G-5‟
Question. What is the role of 'Ori‘ in any plasmid?
Answer: Ori is a nucleotide sequence in plasmid vector which makes any piece of DNA linked to this sequence replicate with in host cells.
Ori also determines the copy number of linked DNA.
Question. Name the commonly used vector for trans formation in plant cell?
Answer: Agrobacterium tumefaciens.
Question. Why is 'Origin of replication‘ (Ori) required to facilitate cloning into a vector?
Answer: Ori is a sequence from where replication starts and any piece of DNA to replicate in the host
cell needs to be linked to it.
*It also controls the copy number of the linked DNA.
Question. The gene of interest is cloned at which position in plasmid PBR322 to facilitate quick selection?
Answer: The gene of interest is inserted at restriction enzyme site located in any one of the antibiotic resistance genes or selectable marker gene. The restriction sites are Hind III, EcoR I, BamH I, Sal I, Pvu I, Pst I, Cla I and the selectable markers are ampR and tetR.
Question. Which enzymes are used for releasing macromolecules (DNA) from cell envelope?
Answer: Lysozyme for bacteria, cellulase for plant cell and chitinase for fungus.
Question. State what happens when an alien gene is ligated at Sal I site of pBR322 plasmid.
Answer: If an alien gene is ligated at Sal I site of tetracycline resistance gene in the vector pBR322, the recombinant plasmid will lose its tetracycline resistance.
Question. Why is Agrobacterium tumefaciens considered as a good cloning vector? Explain.
Answer: i) Agrobacterium tumefaciens
is a soil bacterium which causes diseases in many dicot plants?
(ii)It is able to deliver a piece of DNA known as T-DNA, to transform the normal cells into tumour cells and direct these tumour cells to produce to chemicals required by the pathogen.
Question. What is EcoRI? How does EcoRI differ from an exonuclease?
Answer: EcoRI is an endonuclease restriction enzyme which cut both the stands of palindromic DNA at a specific position of nitrogen base sequence (GAATTC) while exonuclease removes nucleotides from terminals of DNA strands.
Question. Why we prefer the usage of Taq polymerase in PCR technique?
Answer: It is a thermostable enzyme that can withstand high temperature used in the denaturation and separation of DNA strands. Hence, it can be used for a number of cycles in amplification.
Question. Name the host cells in which microinjection technique is used to introduce an alien DNA.
Answer: The microinjection technique to introduce alien DNA is usually carried out in animal cell, i.e. directly into the nucleus.
Question. What are recombinant proteins? How do bioreactors help in their production?
Answer: Any protein encoding gene is expressed in a heterologous host, is called recombinant protein.
Bioreactors help in the production of recombinant proteins on large scale. A bioreactor provides optimal conditions for achieving the desired recombinant protein by biological
methods.
Question. How does a simply stirred tank bioreactor to differ from sparged stirred – tank‘ bioreactor?
Answer: In the simply stirred tank bioreactor the stirrer facilitates the even mixing and the oxygen availability throughout the process, whereas for proper mixing throughout the reactor in the case of sparged stirred-tank bioreactor the air is found to be bubbled.
Question. Name the enzyme used as an alternate selectable marker.
Answer: β --Galactosidase.
Short Answer Type Questions:
Question. What are the two core techniques that enabled the birth of biotechnology?
Answer: The two core techniques that enabled the birth of modern biotechnology are:
Genetic engineering techniques to alter the chemistry of genetic material (DNA and RNA), so that it can be introduced to host organisms to change the phenotype of the host organism.
Maintenance of sterile ambience in chemical engineering processes to enable the growth of only the desired microbe/eukaryotic cell in large quantities for the manufacture of biotechnological products like antibiotics, vaccines, enzymes, etc.
Question. How do bioreactors help in the production of recombinant proteins?
Answer: Small volume cultures cannot yield appreciable quantities of products. To produce these products in large quantities the development of 'bioreactors‘ was required where large volumes of (100-1000 L) of cultures can be processed. Thus, bioreactors can be thought of as vessels in which raw materials are biologically converted into specific products, using
microbial, plant, animal, or human cells or individual enzymes. A bioreactor provides the optimal conditions for achieving the desired product by providing optimum growth conditions like temperature, pH, substrate, salts, vitamins and oxygen.
Question. How are DNA segments separated by gel electrophoresis be visualised and isolated?
Answer: The separated DNA fragments are visualised after staining with ethidium bromide followed by exposure to UV light.
-The bright orange-coloured bands of DNA appears when ethidium bromide-stained gel is exposed to UV light.
-The separated bands of DNA are cut out from the gel piece and extracted from the gel piece.
This is known as elution.
Question. Explain the work carried out by Cohen and Boyer that contributed immensely to biotechnology.
Answer: They isolated antibiotic resistance gene by cutting out a piece of DNA from the plasmid.
The cutting of DNA at specific locations was done with the help of restriction enzymes popularly called as „molecular scissors.
The cut piece of DNA with antibiotic resistance gene was then linked with the plasmid DNA of Salmonella typhimurium acting as vector with the help of the enzyme DNA ligase.
This new autonomously replicating DNA created in vitro with linked fragment of antibiotic resistant gene is called recombinant DNA Recombinant DNA was then transferred into Escherichia coli, where it could replicate using the new host‟s DNA polymerase enzyme. The ability to multiply copies of antibiotic resistance gene in E. coli was called cloning of antibiotic resistance gene in E. coli.
Question. List 6 recombinant proteins which are used in medical practice? Find where they are used as therapeutics.
Answer: RECOMBINANT PROTEIN THERAPEUTIC USE
1. INSULIN For treatment of diabetes mellitus
2.HUMAN GROWTH HORMONE For treatment of dwarfism
3. INTERFERONS For the treatment of viral diseases, cancer and AIDS
4. STREPTOKINASE For treating thrombosis
5. INTERLEUKINS For treating sepsis and cancer
6. HEPATITIS -B SURFACE ANTIGEN Vaccine against hepatitis B
Question. What are the components of a bioreactor?
Answer: The bioreactor has the following components:
• An agitator system.
• An oxygen delivery system.
• A foam control system.
• A temperature control system.
• pH control system and
• Sampling ports.
Question. How are restriction endonuclease enzymes named? Write examples.
Answer: The naming of restriction enzymes is as follows:
The first letter of the name comes from the genus and the next two letters from the name of the species of the prokaryotic cell from which they are isolated.
The next letter comes from the strain of the prokaryote.
The roman numbers following these four letters indicate the order in which the enzymes were isolated from that strain of the bacterium.
eg.EcoR I is isolated from Escherichia coli RY.
Question. Besides better aeration and mixing properties, what other advantages do stirred tank bioreactors have over shake flasks?
Answer: Shake flasks are used for growing and mixing the desired materials on a small scale in the laboratory. A large scale production of desired biotechnological product is done by using ‘bioreactors’. Besides better aeration and mixing properties, the bioreactors have following advantages
(i) Small volumes of cultures are periodically withdrawn from die reactor for sampling.
(ii) It has a foam control system, pH control system and temperature control system.
(iii) Facilitates even mixing and oxygen availability throughout the bioreactor.
Important Notes for Class 12 Biology Chapter 11 Biotechnology Principles and Processes
Biotechnology is the technique of using living organisms or enzymes from organisms to produce products and processes useful to humans. The term was coined by Karl Ereky in
1917.
Biotechnology deals with the large scale production and marketing of products such as enzymes, insulin or antibiotics,etc., that are of importance to mankind.
The European Federation of Biotechnology (EFB) has given a definition of biotechnology as the integration of natural science and organisms, cells, parts thereof and molecular analogues for products and services’.
Principles of Biotechnology
Following two core techniques gave birth to modern biotechnology
1. Genetic engineering It is the alteration of the chemistry of genetic material (DNA/RNA) and the introduction of these into host organisms to consequently change in the phenotype of the host organism. Paul Berg is known as the Father of Genetic Engineering.
2. Bioprocess engineering It is the maintenance of sterile conditions in order to enable the growth of only desired microbes or eukaryotic cells in large quantities for the production of antibiotics, enzymes, hormones, vaccines, etc.
Genetic Engineering
- It is the deliberate modification of an organism’s DNA, using various techniques. This altered DNA (recombinant DNA) is then introduced into host organisms to change their phenotype.
- This is followed by growing this genetically modified cell in large quantities, by maintaining sterile environment, for the manufacture of biotechnological products like antibiotics, vaccines, enzymes, etc.
- Techniques of genetic engineering include construction of recombinant DNA, gene cloning and gene transfer. These techniques allow the isolation and introduction of a set of desirable genes without introducing undesirable genes into the target organism.
- Origin of replication (ori) is a specific DNA sequence in the chromosome which can initiate DNA replication. The foreign DNA introduced into the host genome has to be linked with the origin of replication in the host chromosome for the gene to be able to multiply. This is also known as cloning which involves making multiple identical copies of any template DNA. If the foreign gene is not linked to the ori sequence, it may not be able to multiply.l In 1972, the first recombinant DNA was constructed by Stanley Cohen and Herbert Boyer. They isolated the antibiotic resistance gene by cutting out a piece of DNA from a plasmid (autonomously replicating circular extrachromosomal DNA) of Salmonella typhimurium. The cutting of DNA at specific locations became possible with the help of restriction enzymes (molecular scissors).
- The cut pieces of DNA were then linked with the plasmid DNA using DNA ligase enzyme. These plasmids act as vectors to transfer the piece of DNA attached to it into the host organism. This makes a new circular autonomously replicating DNA created in vitro and is known as the recombinant DNA.
- The recombinant DNA is transferred into Escherichia coli, a bacterium closely related to Salmonella, where it replicates using the new host’s DNA polymerase enzyme and makes multiple copies of itself. It also produces multiple copies of the antibiotic resistance gene in the new host (E. coli). This process is called as cloning of antibiotic resistance gene in E. coli.
- There are three basic steps involved in genetically modifying an organism
– Identification of DNA with desirable genes.
– Introduction of the identified DNA into the host.
– Maintenance of introduced DNA in the host and
transfer of the DNA to its progeny.
Tools of Recombinant DNA Technology
Genetic engineering or recombinant DNA technology can be accomplished only with the usage of key tools like
(i) Enzymes
(ii) Cloning vectors
(iii) Competent host organisms
These tools of rDNA technology are discussed in details below
Enzymes
There are various enzymes involved in genetic engineering which simplifies this complex process.
The enzymes involved are
1. Restriction Enzymes (Molecular scissors)
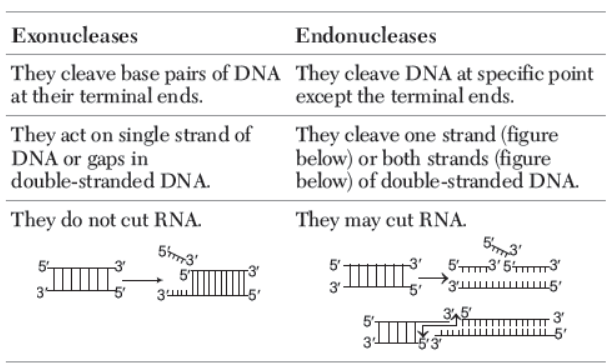
- These are enzymes which are used for cutting of DNA at specific locations during rDNA technology. These enzymes belong to a larger class of nucleases, which are of two types
(i) Exonucleases remove nucleotides from the ends of the DNA (either 5¢ or 3¢) in one strand of duplex.
(ii) Endonucleases make cuts at specific position within DNA.
- The first restriction endonuclease named Hind II was isolated by Smith Wilcox and Kelley in 1968 . It was found that it always cuts DNA molecule at a particular point recognising a specific sequence of the six base pairs known as the recognition sequence for Hind II.
- More than 900 restriction enzymes are known today that have been isolated from over 230 bacterial strains and each of which recognises different recognition sequences. Restriction endonucleases are not present in eukaryotic cells.
Differences between Exonucleases and Endonucleases
Naming of Restriction Enzymes
- The convention for naming these enzymes proceeds in a way that the first letter of the name comes from the genus and the second two letters come from the species of prokaryotic cell from which they were isolated, e.g. EcoRI comes from E. coli RY13.
- The letter ‘R’ is derived from the name of strain.
Roman numbers following the names, indicate the order in which the enzymes were isolated from that strain of bacteria.
Mechanism of Action of Restriction Endonuclease
- Each restriction endonuclease functions by ‘inspecting’ the length of a DNA sequence.
- Once it finds specific recognition sequence, it will bind to DNA and cut each of the two strands of double helix at specific points in their sugar-phosphate backbones.
- Each restriction endonuclease recongnises a specific palindromic nucleotide sequences in the DNA. Palindromes are group of letters that form the same words when read both from forward and backward, e.g. ‘MALAYALAM’. Therefore, the palindrome in DNA is a base pair sequence that is same when read forward or background, e.g. the following sequences read the same on the two strands in 5¢ ¾¾®3¢ direction as well as 3¢ ¾¾®5¢ direction.
5'—— GAATTC——3'
3'—— CTTAAG——5'
- Restriction enzymes cut the strand of DNA a little away from the centre of the palindrome sites, but between the same two bases on the opposite strands
- This leaves single-stranded portion at the ends. These are overhanging stretches called sticky ends present on each strand.
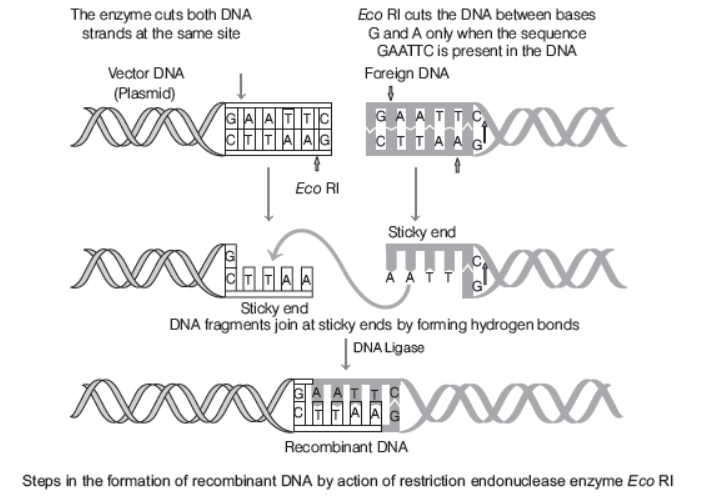
- These are named so because they form hydrogen bonds with their complementary cut counter parts. This stickiness of the ends facilitates the action of the enzyme DNA ligase.
- In genetic engineering, restriction endonucleases are used to form recombinant molecules of DNA, which are composed of DNA from different sources or genomes.
- In order to create a recombinant vector molecule, it is necessary that the vector and the source DNA are cut with the same restriction enzyme, so that the resultant DNA fragments have the same sticky ends, which are complementary to each other and can be joined together
(end-to-end) using DNA ligases.
2. DNA Ligases (Molecular glue)
These enzymes repair broken DNA by joining two nucleotides. They are used in genetic engineering to reverse the action of restriction enzymes, i.e. to join complementary DNA strands/ends together, e.g. T4 DNA ligase.
3. Alkaline Phosphatase (AP)
This enzyme removes the phosphate group from the 5´ end of a DNA molecule, leaving a free 5´ hydroxyl group, it prevents unwanted self-ligation of vector DNA molecules during the formation of recombinant DNA.
4. DNA Polymerases
This enzyme helps in in vitro synthesis of complementary DNA (cDNA) strand on DNA templates.
Cloning Vectors
The DNA molecule that can carry a foreign DNA segment and replicate inside the host cell is called as a vector.
- Plasmids and bacteriophages are used as cloning vectors.This is because plasmids and bacteriophages have the ability to replicate within the bacterial cell independent of chromosomal DNA.
- Bacteriophages have very high copy numbers of their genome within the bacterial cells. But in case of plasmids, some may have only one or two copies per cell whereas others may have 15-100 copies per cell.
- Cloning vectors use the machinery of bacterial cell to replicate and thereby, increase the copy number (make clones) of the DNA inserted into them.
- These help easy linking of foreign DNA and allow the selection of recombinants (bacterial cells that have picked up recombinant plasmid) from non-recombinants (those who have not).
Features of Cloning Vector
All vectors have four special features that are required to facilitate cloning into a vector.
1. Origin of replication (ori) is the sequence from where replication starts and any piece of DNA when linked to this sequence can be made to replicate within the host cells. This sequence also controls the copy number of the linked DNA.
2. Selectable marker helps in identifying and eliminating non-transformants and selectively permitting the growth of the transformants.
Transformation is the procedure through which a piece of DNA is introduced into a host bacterium. Normally, antibiotics such as ampicillin, chloramphenicol, tetracycline or kanamycin, etc., are considered useful selectable markers.
3. Cloning (recognition) sites These are generally required to link the foreign or alien DNA with the vector. For this, the vector requires very few or single recognition sites for commonly used restriction enzymes. If more than one recognition site is present within the vector, it will generate several fragments that will lead to more complication in gene cloning.
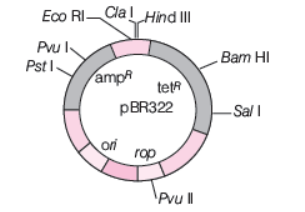
E. coli cloning vector pBR322 showing restriction sites (Hind III, EcoRI, BamHI, Sal I, Pvu II, Pst I, Cla I), ori and antibiotic resistance genes (ampR and tetR). rop codes for the proteins involved in the replication of the plasmid
The ligation of the foreign DNA is carried out at a restriction site present in one of the two antibiotic resistance genes. Selection of recombinants formed can be done by one of the following methods, given below
(i) Inactivation of antibiotic resistance gene If a foreign DNA is ligated at the BamHI site of tetracycline-resistance gene in the vector pBR322, the recombinant plasmid will loose tetracycline resistance.
But, it can still be selected out from nonrecombinant ones by plating transformants on ampicillin containing medium. The transformants growing on medium containing ampicillin are then transferred on a medium containing tetracycline. The recombinants will grow on ampicillin containing medium, but not on that containing tetracycline. On the other side, non-recombinants will grow on both media and thus, recombinants are selected.
In this example, one antibiotic resistance gene helps in selecting the transformants, whereas the other antibiotic resistance gene gets inactivated due to insertion of alien DNA and helps in selection of recombinants.
(ii) Insertional inactivation The selection of recombinants due to inactivation of antibiotic gene is a cumbersome procedure, because it requires simultaneous plating on two plates having different antibiotics. Therefore, the alternative selectable markers have been developed, which differentiate recombinants from non-recombinants. It is based on their ability to produce colour in the presence of chromogenic substrate.
In this process, a recombinant DNA is inserted within the coding sequence of an enzyme b-galactosidase. This results into inactivation of the enzyme, which is referred to as insertional inactivation. Therefore, the bacterial colonies having inserted plasmid, shows no colouration (recombinant colonies), while those without plasmid will show blue colour. Here, b-galactosidase works as a reporter enzyme (produced by reporter gene) whose inactivity helps to identify transformed and non-transformed bacterial colonies.
4. Vectors for cloning genes in plants and animals A pathogen of several dicot plants is able to deliver a piece of DNA, i.e. ‘T-DNA’ to transform normal plant cells into a tumour cells and disect these tumour cells to produce the chemicals required by the pathogen. Similarly, retrovirus in animals have the ability to transform normal cells into cancerous cells.
- The tumour inducing (Ti) plasmid of Agrobacterium tumefaciens has now been modified into a cloning vector which is able to use the mechanisms to deliver genes of our interest into a variety of plants.
- Similarly, retroviruses can be disarmed and used to deliver desirable genes into animal cells.
Competent Host Organisms
DNA is a hydrophilic molecule, so it cannot pass through cell membranes. In order to force bacteria to take up the plasmid, the bacterial cells must first be made competent. The competency is the ability of a cell to take up foreign DNA.
The cells can be made competent by following methods
1. Chemical method (CaCl 2 method) In this, the cells are treated with a specific concentration of a divalent cation such as calcium which increases the efficiency with which DNA enters the bacterium through the pores in its cell wall. The cells are incubated with recombinant DNA on ice, followed by placing them briefly at 42°C and then putting them back on ice. This treatment that enables the bacteria to take up the recombinant DNA is known as heat shock method.
2. Physical method The microinjection is the physical method used to introduce foreign DNA into host cell. In this method, recombinant DNA is directly injected into the nucleus of an animal cell.
Biolistics or Gene gun is another method suitable for introduction of DNA into plants, in which cells are bombarded with high velocity microparticles of gold or tungsten coated with DNA.
3. Disarmed pathogen vectors In this method, vectors that can cause infection are used to infect the cell to transfer the recombinant DNA into the host, e.g. retrovirus, papilloma virus and adenovirus in case of animals and Agrobacterium in case of plants.
Processes of Recombinant DNA Technology
Recombinant DNA technology involves various steps in a specific sequence such as isolation of the desired genetic material (DNA), cutting of DNA at specific locations, isolation of desired DNA fragment, amplification of gene of interest by PCR, ligation of DNA fragments into a vector, insertion of recombinant DNA into host cell, culturing the host cells in a medium at large scale and extraction of the desired product.
Isolation of the Genetic Material (DNA)
Nucleic acid is the genetic material, which is present in all living organisms. In majority of organisms, this is present in the form of Deoxyribonucleic Acid (DNA), which must be present in pure form, i.e. free from other macromolecules in order to cut with restriction enzymes.
This is done in the following sequence
(i) Since DNA is enclosed within membranes, so in order to release DNA along with other macromolecules such as proteins, polysaccharides and lipids, bacterial cells, plant or animal tissues are treated with the enzyme such as lysozyme (bacteria), cellulase (plant cells), chitinase (fungus), respectively.
(ii) RNA can be removed by treating it with ribonuclease,whereas proteins can be removed by its treatment with protease.
(iii) Other molecules can be removed by appropriate treatments and ultimately purified DNA will precipitate out, after the addition of chilled ethanol.
This can be seen as collection of fine threads in the suspension which can be removed by spooling.
Cutting of DNA at Specific Locations
Restriction enzyme digestions are performed by incubating purified DNA molecules with the restriction enzyme. This is done at the optimal conditions for that specific enzyme.
Separation and Isolation of DNA Fragments
- The cutting of DNA by restriction endonucleases results in the fragments of DNA which are separated by a technique known as gel electrophoresis.
- Since, the DNA fragments are negatively charged molecules,so they can be separated by applying an electric field which makes DNA molecules move towards the anode (+), under an electric field through an appropriate medium or matrix.
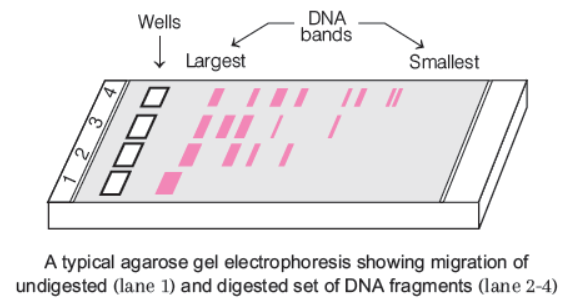
A typical agarose gel electrophoresis showing migration of undigested (lane 1) and digested set of DNA fragments (lane 2-4)
- The most commonly used matrix is agarose, a natural polymer extracted from sea weeds. DNA fragments resolve (separate) according to their size through sieving effect provided by the agarose gel.
- Hence the smaller the fragment size, farther it moves. The separated DNA fragments can be visualised only after staining the DNA with Ethidium Bromide (EtBr) followed by exposure to UV radiation. The bright orange coloured bands of DNA, are seen when exposed to UV light.
- These separate stained bands of DNA are cut out from the agarose gel and extracted from the gel piece, by a process known as elution. Thus, the DNA fragments (containing gene of interest) are purified and used for the construction of recombinant DNA by joining them with cloning vectors in the presence of DNA ligase.
Amplification of Gene of Interest Using PCR
PCR stands for Polymerase Chain Reaction, a method of amplifying the fragments of DNA. This method can make multiple copies of even a single DNA fragment or the gene of interest in a test tube. The reaction mixture requires
(i) DNA template The double-stranded DNA that needs to be amplified.
(ii) Primers These are chemically synthesised oligonucleotides (short segment of DNA) that are complementary to the regions of DNA template.
(iii) Enzymes Two commonly used enzymes in PCR reaction are
- Taq polymerase It is isolated from a thermophilic bacterium, i.e. Thermus aquaticus and has a property to remain active during the high temperature induced denaturation of double-stranded DNA. It also helps in the amplification of a segment of DNA.
- Vent polymerase It is isolated from Thermococcus litoralis.
(iv) Nucleotide bases These are added by DNA polymerase to the growing chain.
- Three main steps involved in the PCR technique are
Step I Denaturation The double-stranded DNA is denatured by using high temperature of 95° C for 15 seconds. Now each separated single strand acts as a template for DNA synthesis.
Step II Annealing Two sets of oligonucleotide primers are annealed (hybridised) to the separated single-strands. This step is carried out at a slightly lower temperature (40-60° C).
Step III Extension The thermostable enzyme Taq DNA polymerase is used in this reaction, extends the primers by adding dNTPs (deoxynucleoside triphosphates) complementary to those of the template DNA. Mg2+ is required as a cofactor for thermostable DNA polymerase.
- These steps are repeated many times in order to obtain several copies of desired DNA
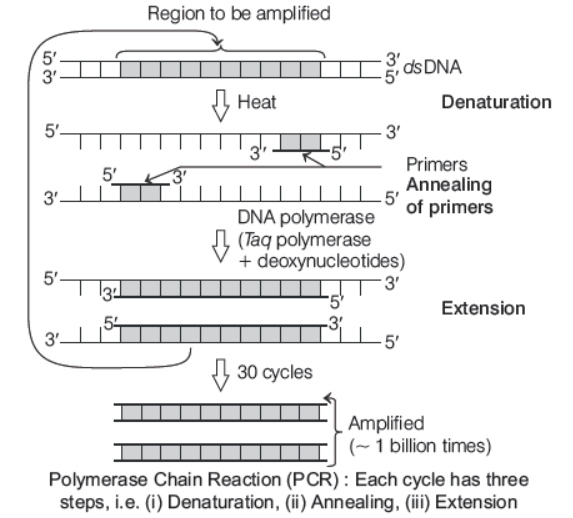
- Applications of PCR are in rDNA technology, DNA sequencing, DNA fingerprinting, early diagnosis of infections, disease, etc.
Ligation of DNA Fragment into a Vector
This process requires a vector DNA and a source DNA. In order to obtain sticky ends, both of these should cut with the same endonuclease, after which, both are ligated by mixing vector DNA, gene of interest and enzyme, DNA ligase to form the recombinant DNA/hybrid DNA.
Insertion of Recombinant DNA into the Host Cell/Organism
This can occur by several methods, before which the recipient cells are made competent to receive the DNA. If a recombinant DNA bearing gene for resistance to an antibiotic (e.g. ampicillin) is transferred into E. coli cells, the host cells become transformed into ampicillin resistant cells. The ampicillin resistance gene in this case is called a selectable marker. When transformed cells are grown on agar plates containing ampicillin, only transformants will grow and others will die.
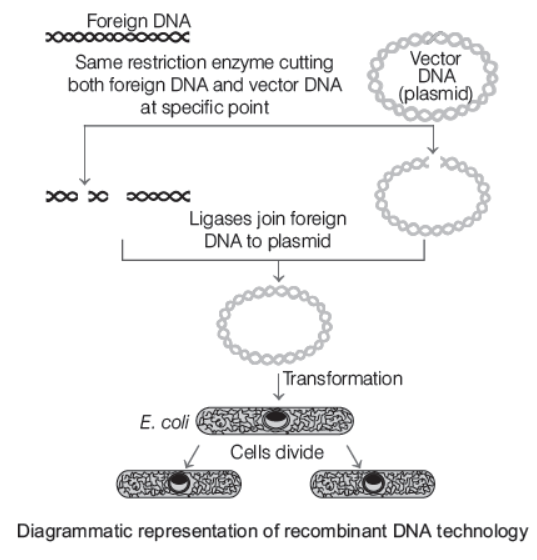
Obtaining or Culturing the Foreign Gene Product
Any protein encoding gene expressed in a heterologous host, it is called recombinant protein. The cells harbouring cloned genes of interest are grown on a small scale in the laboratory (i.e. appropriate nutrient medium at optimal conditions). These cell cultures are used for extracting the desired protein using various separation techniques.
Bioreactors
These are the large volume vessels approximately (100-1000 L) in which raw materials are biologically converted into specific products, individual enzymes, etc., using microbial, plant, animal or human cells.
A bioreactor provides the optimal conditions for achieving the desired product by providing optimum growth conditions like temperature, pH, substrate, salts, vitamins and oxygen.
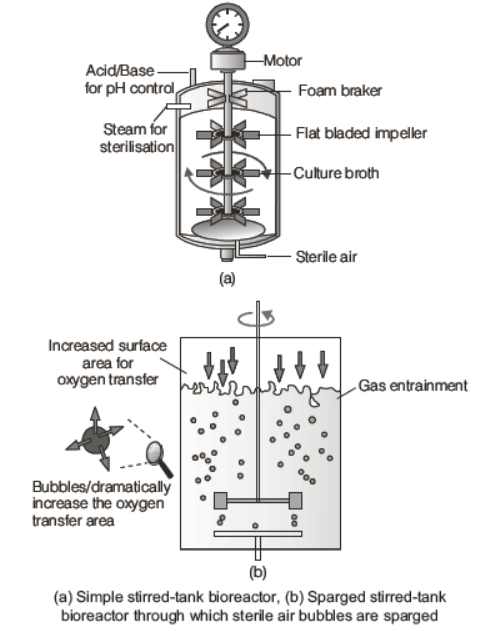
The cells can also be multiplied in a continuous culture system. In this, the used medium is drained out from one side while fresh medium is added from the other side to maintain the cells in their physiologically most active log/exponential phase. This type of culturing method produces a larger biomass leading to higher yields of desired protein.
The components of a bioreactor are
(i) An agitator system
(ii) An oxygen delivery system
(iii) A foam control system
(iv) A temperature control system
(v) pH control system
(vi) A sampling port to withdraw culture periodically
The most commonly used bioreactors are of stirring type. Stirring type bioreactors are further of two types, i.e. simple and sparged.
- A simple stirred-tank bioreactor is usually cylindrical or with a curved base to facilitate even mixing of reactor contents.
- The sparged-stirred-tank bioreactor also facilitates the mixing of components and ensures oxygen availability throughout the bioreactor. Alternatively, air is bubbled through the reactor. Which sterile air bubbles are sparged.
Downstream Processing
After completion of the biosynthetic phase, the product has to be subjected through a series of processes before it is ready for marketing as a finished product. The processes include (i) separation and (ii) purification of products, which are collectively called the downstream processing.
The product is then subjected to quality control testing and formulated with addition of kept in suitable preservatives.
Downstream processing and quality control test generally differ from product to product.
| CBSE Class 12 Biology Organisms And Populations Worksheet Set A |
| CBSE Class 12 Biology Organisms And Populations Worksheet Set B |
| CBSE Class 12 Biology Ecosystem Worksheet Set A |
| CBSE Class 12 Biology Ecosystem Worksheet Set B |
| CBSE Class 12 Biology Ecosystem Worksheet Set C |
Worksheet for CBSE Biology Class 12 Chapter 9 Biotechnology Principles and Processes
We hope students liked the above worksheet for Chapter 9 Biotechnology Principles and Processes designed as per the latest syllabus for Class 12 Biology released by CBSE. Students of Class 12 should download in Pdf format and practice the questions and solutions given in the above worksheet for Class 12 Biology on a daily basis. All the latest worksheets with answers have been developed for Biology by referring to the most important and regularly asked topics that the students should learn and practice to get better scores in their class tests and examinations. Expert teachers of studiestoday have referred to the NCERT book for Class 12 Biology to develop the Biology Class 12 worksheet. After solving the questions given in the worksheet which have been developed as per the latest course books also refer to the NCERT solutions for Class 12 Biology designed by our teachers. We have also provided a lot of MCQ questions for Class 12 Biology in the worksheet so that you can solve questions relating to all topics given in each chapter.
You can download the CBSE Printable worksheets for Class 12 Biology Chapter 9 Biotechnology Principles and Processes for latest session from StudiesToday.com
There is no charge for the Printable worksheets for Class 12 CBSE Biology Chapter 9 Biotechnology Principles and Processes you can download everything free
Yes, studiestoday.com provides all latest NCERT Chapter 9 Biotechnology Principles and Processes Class 12 Biology test sheets with answers based on the latest books for the current academic session
CBSE Class 12 Biology Chapter 9 Biotechnology Principles and Processes worksheets cover all topics as per the latest syllabus for current academic year.
Regular practice with Class 12 Biology worksheets can help you understand all concepts better, you can identify weak areas, and improve your speed and accuracy.

